

 |
GPI Lipid Anchor Project |  |


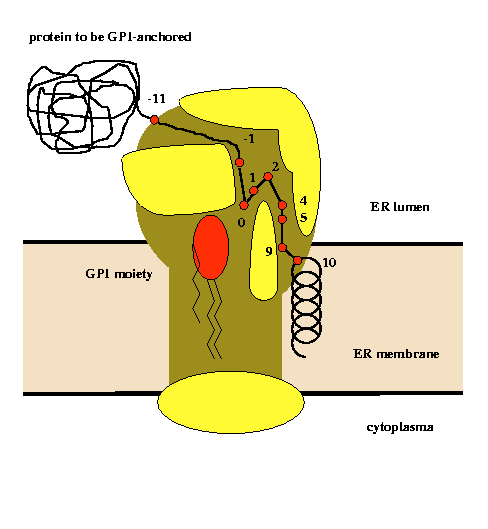
Glycosylphosphatidylinositol (GPI) lipid anchoring is a common posttranslational modification known mainly from extracellular eukaryotic proteins. Attachment of the GPI moiety to the carboxyl terminus (omega-site) of the polypeptide follows after proteolytic cleavage of a C-terminal propeptide. For the first time, a new prediction technique locating potential GPI-modification sites in precursor sequences has been applied for large-scale protein sequence database searches. The composite prediction function (with separate parametrisation for metazoan and protozoan proteins) consists of terms evaluating both amino acid type preferences at sequence positions near a supposed omega-site as well as the concordance with general physical properties encoded in multi-residue correlation within the motif sequence. The latter terms are especially successful in rejecting non-appropriate sequences from consideration. The algorithm has been validated with a self-consistency and two jackknife tests for the learning set of fully annotated sequences from the SWISS-PROT database as well as with a newly created database "big-PI" (more than 300 GPI-motif mutations extracted from original literature sources). The accuracy of predicting the effect of the mutations in the GPI sequence motif was above 83%. Lists of potential precursor proteins which are non-annotated in SWISS-PROT and SPTrEMBL are presented on this WWW-page. The algorithm has been implemented in the prototype software "big-PI predictor" which may find application as a genome annotation and target selection tool.

|
An analysis of physical properties of amino acid residues at given sequence positions in the vicinity of the GPI-modification site allowed the construction of a model of the active site of the putative transamidase complex (Eisenhaber et al., Prot.Eng., 11, 1155-1161,1998). |
 Protozoa
Protozoa Metazoa
Metazoa Protozoa
Protozoa Metazoa
Metazoa Protozoa
Protozoa Metazoa
Metazoa
 Protozoa
Protozoa Metazoa
Metazoa Protozoa
Protozoa Metazoa
Metazoa other
other Protozoa
Protozoa Metazoa
Metazoa other
other

Last modified: 12th June 2002