
Glycogen- and PP1C-binding motifs of PTG (Protein Targeting
to Glycogen) and other protein phosphatases
Peer Bork
Thomas Dandekar
Frank Eisenhaber
Martijn Huynen
REFERENCE:
Bork P., Dandekar Th., Eisenhaber F., Huynen M.
"Characterization of targeting domains by sequence analysis: glycogen-binding domains in protein phosphatases"
Journal of Molecular Medicine, 76 (1998) 77-79
Printen et al. (Science v.275 (1997) pp.1475-1478) have identified a glycogen-sensitive subunit of protein phosphatase 1 (PP1G) that targets proteins to glycogen.
Using sequence analysis methods, we were able to identify
 I. the location of a putative PP1C-binding site (within the first 140 residues of PTG)
as well as
I. the location of a putative PP1C-binding site (within the first 140 residues of PTG)
as well as
 II. the location and domain structure of the glycogen binding site (residues 140-240 of PTG)
II. the location and domain structure of the glycogen binding site (residues 140-240 of PTG)
within the sequence of PTG and other related yeast and animal phosphatases.
The
alignment 5 animal PP1Gs and 4 yeast relatives confines the region of similarity to the central part of PTG that covers only about 100 residues. Subjecting this region to
iterative motif and profile searches, numerous high affinity starch-binding
domains of of distinct glycohydrolases can be retrieved.
The amino acid sequence of the second domain is related to the macromolecular carbohydrate (glycogen, starch, etc.) binding domain of AMYG_RHIOR and many other amylases, cyclodextrinases, starch synthetases and the like. The 3D-structure of this domain is known as a beta-sandwich (similar structures are 1kul, 1pam, 1cgt, 1cdg or 1cyg).
Supplementary information:
Fig. 1 The docking surface of the glycogen-binding region (motif 2)
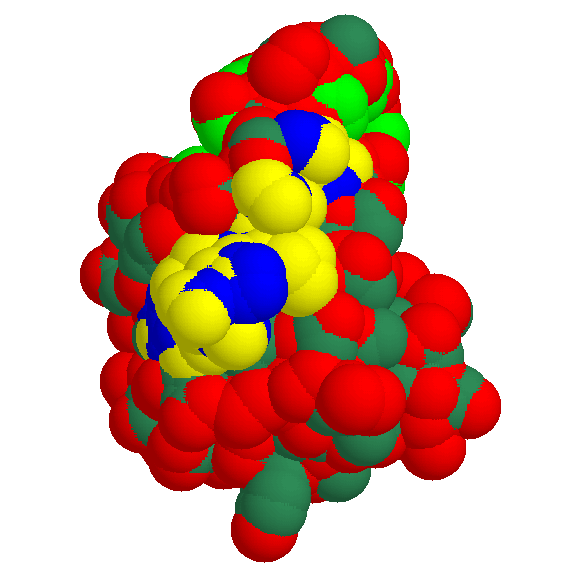
We present the bulk-water accessible surface of 1kul, the granular starch-binding domain of glucoamylase from Aspergillus niger as it is recognized by other molecules in a docking act. The globule is shown from the side of motif 2.
Mapping the conserved residues onto the predicted structure reveals the exposed polar amino acids that are the likely candidates for the direct
interaction with glycogen.
- yellow - polar atoms of residues 589-599 in 1kul (motif 2)
- blue - non-polar atoms of residues 589-599 in 1kul (motif 2)
- red - other polar atoms
- greenblue - other non-polar atoms
- green - non-polar atoms of largest hydrophobic patch
Fig. 2 Secondary structure and side chains of the glycogen-binding domain
(motif 2)
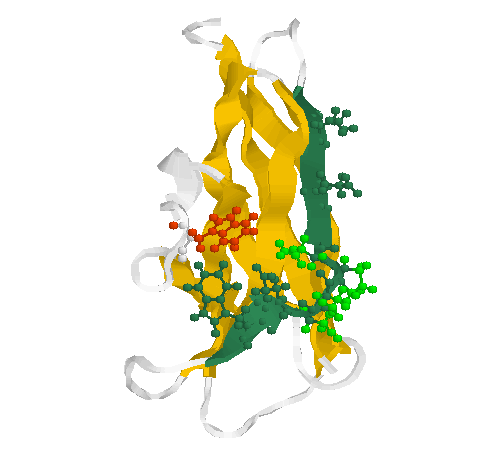
The motif 2 (residues Glu589-Thr599 in 1kul) is shown in greenblue with all side chains pointing outwards in sticks & balls representation. The side chains of the loop (residues Ser592-Asn595) are coloured green. Two conserved aromates (here tryptophanes Trp543 (red) and Trp 590 (greenblue)) play obviously a structural role forming a contact inside the globule.
Fig. 3 Structure of the glycogen-binding domain
(motif 1)
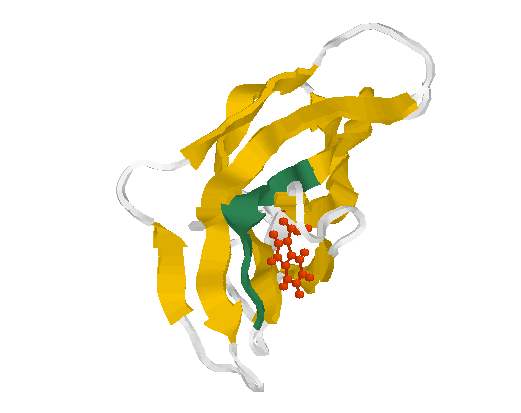
The motif 1 (residues Phe519-Thr526 in 1kul) is shown in greenblue. The conserved aromate (here tryptophane Trp563 (red)) plays obviously a structural role forming a contact inside the globule.

Last modified: Feb 11 2000



 I. the location of a putative PP1C-binding site (within the first 140 residues of PTG)
as well as
I. the location of a putative PP1C-binding site (within the first 140 residues of PTG)
as well as
 II. the location and domain structure of the glycogen binding site (residues 140-240 of PTG)
II. the location and domain structure of the glycogen binding site (residues 140-240 of PTG)



