
MyristoylCoA:Protein N-Myristoyltransferase
Supplementary material to
N-terminal N-Myristoylation of Proteins:
I. Refinement of the Sequence Motif and its Taxon-specific Differences
II. Prediction of Substrate Proteins from Amino Acid Sequence
 |
NMT MyristoylCoA:Protein N-Myristoyltransferase Supplementary material to N-terminal N-Myristoylation of Proteins: I. Refinement of the Sequence Motif and its Taxon-specific Differences II. Prediction of Substrate Proteins from Amino Acid Sequence |
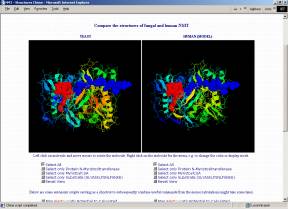
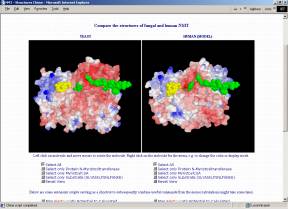
Crystal structures of NMT have been derived for Candida albicans (1NMT, Weston et al. 1998) and Saccharomyces cerevisiae (2NMT, Bhatnagar et al. 1998; 1IID,1IIC, Farazi et al. 2001). However, the high sequence similarity of at least 40% allows homology modeling for the NMTs of many other organisms. The structure of the substrate binding pockets is of major interest because selective inhibition of a pathogenic organism's NMT (without affecting the human one) seems to open completely new ways for medical treatments.
Would you like to see NMT from the view of a
substrate ?
Then I may invite you to a virtual journey
through the NMT binding pocket (download MPEG-movie
[2.5 MB]).
You can also download either our pdb-files directly or RASMOL-scripts to load the molecules in a predefined, recommended view:
NMT_YEAST.pdb (PDB entry 1IID (Farazi et al. 2001) with an elongated substrate peptide)
NMT_YEAST.rasmol (corresponding RASMOL script file, pdb coordinates included; start with 'rasmol -script NMT_YEAST.rasmol')
NMT_HUMAN.pdb (model, created by homology modeling with the Swiss-PdbViewer, Guex & Peitsch 1997)
NMT_HUMAN.rasmol (corresponding RASMOL script file, pdb coordinates included; start with 'rasmol -script NMT_HUMAN.rasmol')
Technical details:
The yeast structure is basically PDB entry 1IID, but we elongated the
octapeptide representing the substrate to the undecapeptide GLYASKLFSNL because
we assume that there can also be interaction of substrate positions 7-11 with
the enzyme. However, definitely to a lower extent than with the first 6
residues.
The human NMT (NMT1_HUMAN, P30419) was modeled
according to structures 2NMT (Bhatnagar et al., 1998) and 1IID (Farazi et
al., 2001)
by utilizing the Swiss-PdbViewer modeling environment (Guex & Peitsch, 1997) except for the N-terminal 80 residues which do not
have a counterpart in the yeast structure and which function as targeting signal
for the ribosomal compartment in the long isoform (Glover et al., 1997). Furthermore, 4 regions (Asp67-Ile68,
Ser303-Lys308, Thr355-Lys371 and Gly435-Arg447) that represent external loops in
the Saccharomyces cerevisiae structure are not conserved in the human
sequence. Energy minimization was carried out with the GROMOS96 force
field (Stocker & van Gunsteren, 2000; Parameter file: IFP43B1; Topology file: MTB43B1;
Method one: steepest descent, cycles: 200, constraints: 25/C-factors, conjugate
gradient method, cycles: 300, constraints: 2500/C-factors). RMS deviation of our
model towards the crystal structure 1IID is 0.84 Å for the Cα
atoms over the complete structure.
If you have the CHIME browser plugin (freely available from MDL; unfortunately only for Windows and Macintosh), you can compare the structures directly on the following interactive page that allows you to visualize, freely rotate, zoom, translate,... the fungal and human NMTs and even lets you calculate and map the electrostatic or the molecular lipophilicity potential to the surfaces. Automatic scripts are prepared to facilitate usage and visualization.
ENTER
THE INTERACTIVE NMT-STRUCTURE COMPARISON PAGE
(Requires
the Chime plugin
to be installed)
If you are using Linux, you unfortunately miss the excellent surface visualizations created by chime unless you run a Windows emulator like Win4lin or VMware. It is more difficult to run Chime under UNIX, Sun/Solaris or SGI/Irix because it requires fast network access to a Windows NT server and the expensive software MetaFrame from Citrix to open a remote window (so, does Windows really have one advantage?).