

 |
GPI Lipid Anchor Project |  |


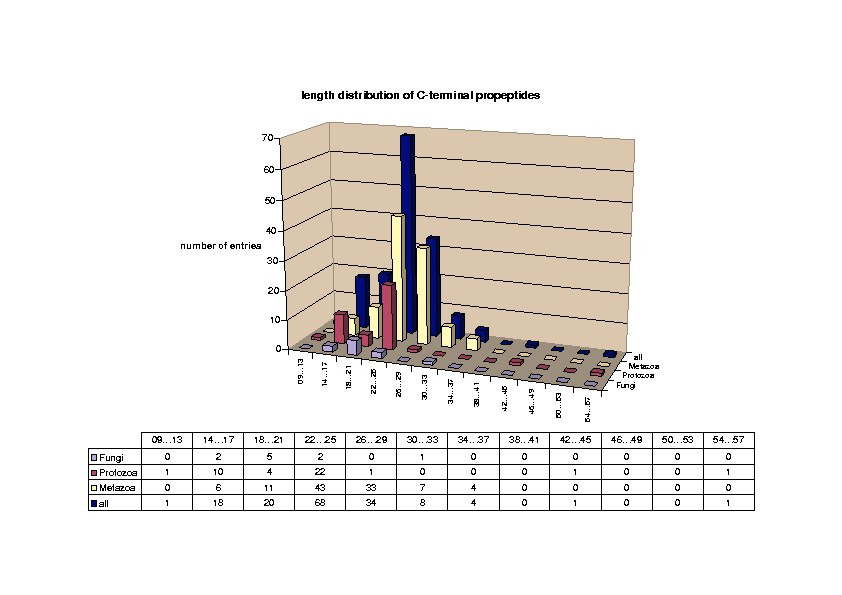
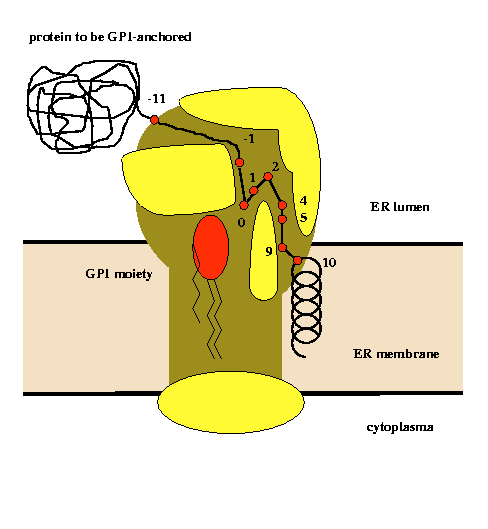
Glycosylphosphatidylinositol (GPI) anchoring is a common posttranslational modification of extracellular eukariotic proteins. Attachment of the GPI moiety to the carboxyl terminus (omega-site) of the polypeptide occurs after proteolytic cleavage of a C-terminal propeptide. In this work, the sequence pattern for GPI-modification has been analyzed in terms of physical amino acid properties based on a database analysis of annotated proprotein sequences. In addition to a refinement of previously described sequence signals, we report conserved sequence properties in the regions (omega-11)...(omega-1) and (omega+4)...(omega+5). We present statistical evidence for volume-compensating residue exchanges with respect to the positions (omega-1)...(omega-2). Differences between protozoan and metazoan GPI-modification motifs consist mainly in variations of preferences to amino acid types at the positions near the omega-site and in the overall motif length. The variations of polypeptide substrates are exploited to suggest a model of the polypeptide binding site of the putative transamidase, the enzyme catalyzing the GPI-modification. The volume of the active site cleft accommodating the four residues (omega-1)...(omega+2) appears about 540 ų.

 List of 155 SWISS-PROT entries with annotated GPI-site
List of 155 SWISS-PROT entries with annotated GPI-site
 amino acid property library
amino acid property library
| Correlation data with amino acid properties | |
|---|---|
| Protozoa | Metazoa |
| 1) for alignment of largest subset | 1) for alignment of largest subset |
| 2) for effective amino acid composition of total alignment | 2) for effective amino acid composition of total alignment |

Last modified: 11th June 2002