


 |
 |
 |
| Home | Initium | Angiogenesis | FAQ |
| TEM domain structure |
|---|
|
|
|
|
| TEM Sequence Analysis |
|---|
| name | from | to | source/E | description |
|---|---|---|---|---|
| unknown | 1 | 250 | predator | The region is a highly helical region, with aa 170-200 forming a coiled-coil structure with very high probability. |
| RhoGAP | 269 | 419 | Pfam E=5.4e-55 | The finger-arginine residue, is positioned at residue 291, and is proposed to be involved in GAP catalysis by stabilizing the transition state and the critical glutamine. The arginine residue is preceded by an aromatic amino acid residue (F) which is proposed to stabilize the adjacent hydrophobic core and balance the orientation of the arginine finger. |
| proline-rich | 526 | 756 | SEG25 | Highly proline-rich regions usually serve as binding sites for Src homology (SH3) domains and other protein modules. These interactions often mediate the generation of dynamic macromolecular signalling complexes. Five proline-rich sequences contain the potential SH3 binding motif pXPpXP (p proline-preferred, X non-conserved residue): PXPXXP: 559-564, 630-635, 739-744, XXPPXP: 543-548, 738-743. |
| 658 | 724 | SAPS | The region is uncharged, particularly the region from 704 to 724. | |
| 818 |
| Remarks |
|---|
ASSESSMENT OF A POTENTIAL TEM28GAP CATALYTIC ACTIVITY: The TEM28 RhoGAP domain shows conservation of several residues with a prospective role GAP activity. The RhoGAP domain has been shown to be fully alpha-helical containing 8 helices of which two adjacent helices (helices B and F) and an adjoining loop (the catalytic A-A1 loop) generate the G-protein binding site. In TEM28, the finger-arginine residue, which is proposed to be involved in GAP catalysis by stabilizing the transition state and the critical glutamine of GTP-binding proteins is positioned at residue 291 in the A-A1 loop. This residue is preceeded by an aromatic amino acid residue (F290) which is proposed to stabilize the adjacent hydrophobic core and balance the orientation of the arginine finger in RhoGAPs. During the transition state of the phosphoryl transfer reaction rearrangment of the G-protein-RhoGAP complex occur:1 ) R291 is relocated such that it can assists in catalysis. 2) Switch I and II of the small G-protein are better ordered which is proposed to assist catalysis. This increased order is created through the interaction of a Tyr in switch I with an conserved Asn of Helix F (N401 in TEM28) and the R291 guanidinium group. Consequently, a catalytic GAP activity is proposed for TEM28.
ASSESSMENT OF A POTENTIAL TEM28GAP BINDING ACTIVITY: The G-protein binding B and F helix can be located to the TEM28 regions 321-334 and 398-411 by superposition of the GAP domain with the respective p50RhoGAP domain. The proposed B and F helices are contained in parts of the GAP domain that show highest conservation (identity 92-94%) to nedrin GAP domain and 3BP-1s GAP domain (both identical in the restricted regions) which have been shown to stimulate the intrinsic GTPase activity of RhoA, Cdc42, Rac1 and Rac1, Cdc42 respectively. The specificity of the small GTPase binding activity of TEM28 cannot be predicted.
| Sequence Information |
|---|
| TEM | TEM28 | ||||||||
Reliability of Gen2Tag mapping |
|
||||||||
| Analysis |
|
||||||||
| Protein Description |
|
||||||||
| Protein Sequence |
MKKQFNRMRQLANQTVGRAEKTEVLSEDLLQVEKRLELVKQVSHSTHKKLTACLQGQQGAEADKRSKKLP LTTLAQCLMEGSAILGDDTLLGKMLKLCGETEDKLAQELIHFELQVERDVIEPLFLLAEVEIPNIQKQRK HLAKLVLDMDSSRTRWQQTSKSSGLSSSLQPAGAKADALREEMEEAANRVEICRDQLSADMYSFVAKEID YANYFQTLIEVQAEYHRKSLTLLQAVLPQIKAQQEAWVEKPSFGKPLEEHLTISGREIAFPIEACVTMLL ECGMQEEGLFRVAPSASKLKKLKAALDCCVVDVQEYSADPHAIAGALKSYLRELPEPLMTFELYDEWIQA SNVQEQDKKLQALWNACEKLPKANHNNIRYLIKFLSKLSEYQDVNKMTPSNMAIVLGPNLLWPQAEGNIT EMMTTVSLQIVGIIEPIIQHADWFFPGEIEFNITGNYGSPVHVNHNANYSSMPSPDMDPADRRQPEQARR PLSVATDNMMLEFYKKDGLRKIQSMGVRVMDTNWVARRGSSAGRKVSCAPPSMQPPAPPAELAAPLPSPL PEQPLDSPAAPALSPSGLGLQPGPERTSTTKSKELSPGSAQKGSPGSSQGTACAGTQPGAQPGAQPGASP SPSQPPADQSPHTLRKVSKKLAPIPPKVPFGQPGAMADQSAGQLSPVSLSPTPPSTPSPYGLSYPQGYSL ASGQLSPAAAPPLASPSVFTSTLSKSRPTPKPRQRPTLPPPQPPTVNLSASSPQSTEAPMLDGMSPGESM STDLVHFDIPSIHIELGSTLRLSPLEHMRRHSVTDKRDSEEESESTAL |
||||||||
SAGE-Tag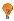 |
CATG-TACAAATCGTT | ||||||||
| EST/cluster Description |
|
||||||||
| EST/cluster Sequence |
GACTGGGAGCAGGCAGCCCGGGCGGAGCGGGCCGGTGCCGAGGACGGCCCCAGGCATTGCTCTGCCCCGG GCATTGCGCGGCGCGCGTGAGGGGGATGCGGCAGGAGGCGGCGCGGCGGGAGGAGTAGGCGGCGGCGCCC TCGGGAGGGAGCTGCGCGCGGGCCAGACGGCGCCCGGAGGCTCCGCAGTGCCGCCGCCGTCGCCCGGGAG GCTCCGCGCGGGAGCCATGTAACCCTGCGGCGGGCTCCGGGCTGCTCCGTCCTTCCCCAGCTCCCGGGCT AGCGCGGCAGCGGGGCCACGATGAAGAAGCAGTTCAATCGCATGCGCCAGCTGGCCAACCAGACGGTGGG CAGGGCTGAAAAGACAGAAGTTTTGAGTGAAGACCTTCTTCAGGTGGAGAAGCGTCTGGAGCTGGTGAAA CAGGTGTCCCACAGCACGCACAAGAAGCTCACCGCATGTCTGCAGGGCCAGCAAGGGGCAGAGGCTGACA AGCGCTCCAAAAAGTTGCCTTTGACAACACTGGCTCAGTGTCTGATGGAGGGGTCAGCTATCCTGGGAGA TGACACACTTCTTGGGAAGATGCTGAAACTCTGTGGAGAGACGGAGGACAAGCTGGCTCAGGAGCTGATA CATTTTGAGTTGCAAGTAGAGAGAGACGTGATTGAGCCCCTGTTTTTGCTGGCGGAGGTGGAAATCCCAA ATATTCAAAAGCAGAGGAAACACTTAGCCAAGTTGGTGCTGGACATGGATTCCTCACGAACCAGGTGGCA GCAGACTTCCAAGTCTTCAGGTTTGTCCAGCAGCTTACAGCCTGCGGGTGCCAAGGCTGATGCCCTCAGG GAAGAAATGGAAGAGGCTGCCAACAGAGTGGAGATTTGCAGGGACCAGCTCTCAGCTGATATGTACAGTT TTGTGGCCAAAGAAATTGACTATGCAAACTACTTTCAAACGCTAATAGAAGTGCAAGCTGAATACCACAG GAAGTCCCTGACACTATTGCAGGCTGTATTGCCTCAGATCAAAGCACAACAGGAGGCCTGGGTAGAGAAG CCTTCCTTCGGGAAGCCGCTGGAGGAGCACCTCACCATCAGCGGCCGGGAGATCGCCTTCCCCATCGAGG CGTGTGTGACCATGCTGCTTGAGTGTGGGATGCAGGAGGAGGGACTCTTCCGAGTAGCCCCCTCTGCCTC CAAACTGAAGAAGCTGAAAGCGGCCCTGGACTGCTGCGTGGTGGATGTGCAGGAGTACTCGGCAGACCCC CACGCAATTGCAGGAGCTTTGAAATCTTACCTCCGAGAGTTGCCAGAACCTCTTATGACCTTTGAACTCT ATGATGAGTGGATCCAGGCTTCCAATGTCCAGGAGCAAGACAAGAAGCTTCAGGCTCTATGGAATGCTTG TGAAAAGTTGCCCAAGGCCAATCACAACAACATCCGATACTTGATAAAATTTTTATCCAAGCTGTCAGAA TATCAAGATGTAAACAAGATGACTCCCAGTAATATGGCAATTGTTTTAGGACCCAACCTCCTATGGCCAC AAGCAGAAGGGAACATTACAGAGATGATGACCACAGTGTCGCTGCAAATTGTTGGGATCATTGAACCTAT CATCCAGCATGCAGACTGGTTCTTCCCTGGGGAGATAGAGTTCAACATTACTGGCAATTATGGGAGTCCA GTACACGTGAACCATAATGCCAACTACAGCTCAATGCCCTCCCCAGACATGGACCCTGCTGACCGGCGCC AGCCCGAGCAGGCCCGCCGGCCCCTCAGCGTCGCCACGGATAATATGATGCTGGAGTTTTACAAAAAGGA TGGCCTTAGGAAAATCCAAAGCATGGGTGTGAGGGTCATGGACACAAACTGGGTGGCTCGAAGAGGCTCC TCGGCCGGTCGGAAAGTGTCCTGCGCCCCGCCCTCCATGCAGCCTCCCGCCCCGCCCGCCGAGCTGGCTG CGCCCCTGCCTTCGCCGCTGCCGGAGCAGCCCCTGGACAGCCCCGCGGCCCCCGCGCTCTCTCCATCCGG CCTGGGCCTCCAGCCTGGGCCCGAGCGCACCAGCACAACAAAAAGCAAGGAACTTTCTCCAGGCTCTGCA CAGAAAGGAAGTCCAGGCTCCAGCCAGGGCACAGCCTGTGCAGGGACTCAACCAGGGGCTCAACCTGGAG CTCAGCCGGGCGCCAGCCCCAGCCCCAGCCAGCCGCCTGCAGACCAGAGTCCTCACACCCTCCGGAAAGT TTCAAAGAAGCTGGCACCGATTCCACCCAAGGTCCCCTTTGGCCAGCCGGGGGCTATGGCAGACCAGTCC GCTGGCCAGCTGTCCCCAGTCAGCCTGTCCCCCACCCCGCCCAGCACCCCGTCACCCTATGGACTGAGCT ACCCTCAGGGGTACTCCTTGGCCTCGGGCCAGCTCTCCCCAGCTGCAGCTCCTCCCCTGGCCTCTCCTTC TGTCTTTACAAGCACTTTGAGCAAATCGCGGCCCACTCCTAAGCCGCGACAGAGACCTACTCTGCCGCCT CCTCAGCCTCCCACAGTAAACCTCTCGGCCTCTAGTCCACAGTCCACGGAGGCCCCCATGCTAGATGGCA TGTCCCCTGGGGAAAGCATGTCTACAGATCTTGTCCACTTTGATATTCCCTCGATCCACATAGAGCTCGG GTCGACGCTCCGCCTGAGTCCCCTGGAGCACATGCGGCGACACTCAGTAACTGACAAGAGGGACTCGGAG GAGGAGTCTGAGAGCACCGCCCTCTGACATGACACCGCCCATCCTGCCTCGCGTGTACATACATCACGGG CCCTAGGAACGCCGCCAGGAGCAGCGTCCATGAGCTTGCCAAGTGTTCTCTGCTGGCTCTTTCCTGCCAC TGCCAACACGAGGTTGGAATTTGGCAGAAAATTGTGATCTCCAGTCCGTGTGGTGATGCTGGTGGTGCAG GTTTTGTTTGTTCCTTTCGGGTGGTGACTTCGGCCTTTTGTTTGACCTTTGCCTTTTGACTTTGTGCCTC TTTTGATCCACTTTCAGCCTCCATGCCAGAAAACACCCACCTCTCCATCCAAGGCTGGTCAGGAACGTCC TTTGCAGGGTCGGGGTGGTGCGGGAGAGGCTCACTTTGCCTGGTTAGACCCAAGGGCTGCTACCTTTTCC TTGGACGGCTCATGTCAGGTCTTGCAGGATCAGTTTAATGGCCACAGAAAGGAAGCAGGACAGCAGGGCC CCTCTCACCCACAACTGGACCAGGTCCAGGATTCTAGCAGTCCTGGGGCACTGACCTTTGCCAGCTACCT GGGGGAGGGCTTGCCACTGGAAAACCTTTCAGGCCGCCCCCATCAGTGGGCTCCAAAGTAAATGGCTGAA AACAAAAATGTTTCACTTCCTAACAGTTTTCCTTTTTCCACTGTGTGACTGAAAGCTCCTATATCATTTT ATATTTCTGAATCTATAAAACAAAACAAACAAGCCTGAAAGTGTCTGGAGGAGCCAAAGGTGGCCTCCCT GTCCCCAAATATATTGGCTATATGAGAGTAATTTTACCCCTCTACGTACCTAAAGGCACCCAGTTCACTA GTCTGTGGGGTCCTGGAGCCTGTCTCTTCTTTCTGGAGGTTCAAACTGAATAGCAATAATTACGTTACCC AAAGCATGTGGAGGAAAAGTGAAACCAGCCACGGAGACGCTGGCCCACGGCTCGGCCTGCGGTGTGGCCT GCTTTGCTCACCAGCGTCAGCCGCTCATTTCCTTCTCATGAAGTCCCATCTGGTCATGGGGACGAGGGCC GGGAGGGCACCGGGTAGCCTTTTCACACTTGGGGATTAGGGGAGTGAGAAAAGATTTGGGCCATGCATGC AAAGTCAAAGTTTAAAATTTTATCCTTTTCAAATAGATGATATAATATACCTATACATGATATAATATTT GTATATATGAAATCTCTCTATATTTGTTTAATTTGAGCCATTCAATCTAAACCAATGTACAGGTGTACAA TGAAAAATTTAAATGCTTAGTTATTTTTCCCAACACAGTGTAAAGTCACCCTCCTCTGAGAGTGGGATGT GCAGAGTTTTGATGTTGCAGCTTTGCTCACTTCCTGGCAAGGGCAGGTCATGCCTCAATTTGTAATGGGA GTCTGGGGTAAGGGTGGGGGTTGAAAGTTGTTATCTTTAAATACATGTACAAATCGTTGTCAAAAGTAAC GTTATTAAAATAGATTTATTATCCCTG |
||||||||
| Genomic Sequence Description |
|
||||||||
| Genomic Sequence |
|
| Literature |
|---|
| PubMed: 10967100 | Nadrin, a novel neuron-specific GTPase-activating protein involved in regulated exocytosis. Harada A, Furuta B, Takeuchi Ki, Itakura M, Takahashi M, Umeda M. J Biol Chem. 2000 Nov 24;275(47):36885-91. |
| PubMed: 10899318 | Functional specificity conferred by the unique plasticity of fully alpha-helical Ras and Rho GAPs. Souchet M, Poupon A, Callebaut I, Leger I, Mornon J, Bril A, Calmels TP. FEBS Lett. 2000 Jul 14;477(1-2):99-105. |
| PubMed: 7526465 | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Science. 1994 Nov 18;266(5188):1241-7. |
| PubMed: 7510218 | Structural basis for the binding of proline-rich peptides to SH3 domains. Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL Cell 1994 Mar 11;76(5):933-45 |
| PubMed: 8438166 | Identification of a ten-amino acid proline-rich SH3 binding site. Ren R, Mayer BJ, Cicchetti P, Baltimore D Science 1993 Feb 19;259(5098):1157-61 |
| Home | Initium | Angiogenesis | FAQ |