
MyristoylCoA:Protein N-Myristoyltransferase
Supplementary material to
N-terminal N-Myristoylation of Proteins:
I. Refinement of the Sequence Motif and its Taxon-specific Differences
II. Prediction of Substrate Proteins from Amino Acid Sequence
 |
NMT MyristoylCoA:Protein N-Myristoyltransferase Supplementary material to N-terminal N-Myristoylation of Proteins: I. Refinement of the Sequence Motif and its Taxon-specific Differences II. Prediction of Substrate Proteins from Amino Acid Sequence |
The variety of organisms revealed by our search makes the NMT an interesting target for broad phylogenetic analysis of eukaryotes in general, since analysis with combined protein data showed striking differences to previous analyses of small ribosomal subunit RNA (Baldauf et al. 2000). As not all full-length protein sequences from the identified organisms are available yet, we had to select a largest subset with homology for each region known to be important for proper functioning of the enzyme.
First,
we concentrated on critical and highly conserved parts of the myristoylCoA
binding pocket in the N-terminal domain of NMT and obtained the following
tree:
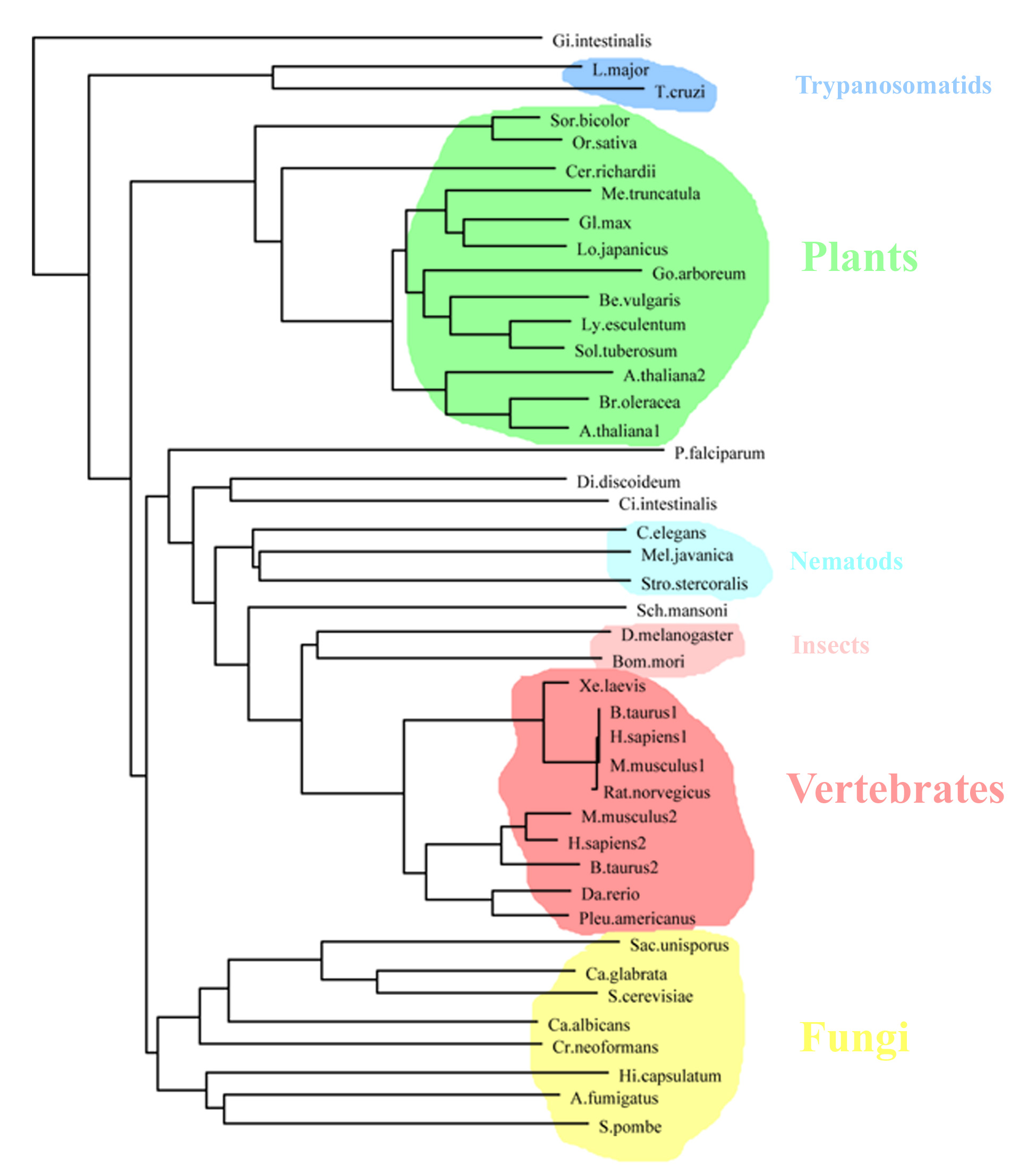
As
can be seen, there are three major separate clusters comprising the
fungal, plant and metazoan sequences.
When
analyzing the C-terminus (catalytic center and part of the substrate
protein binding pocket) we obtained similar results:
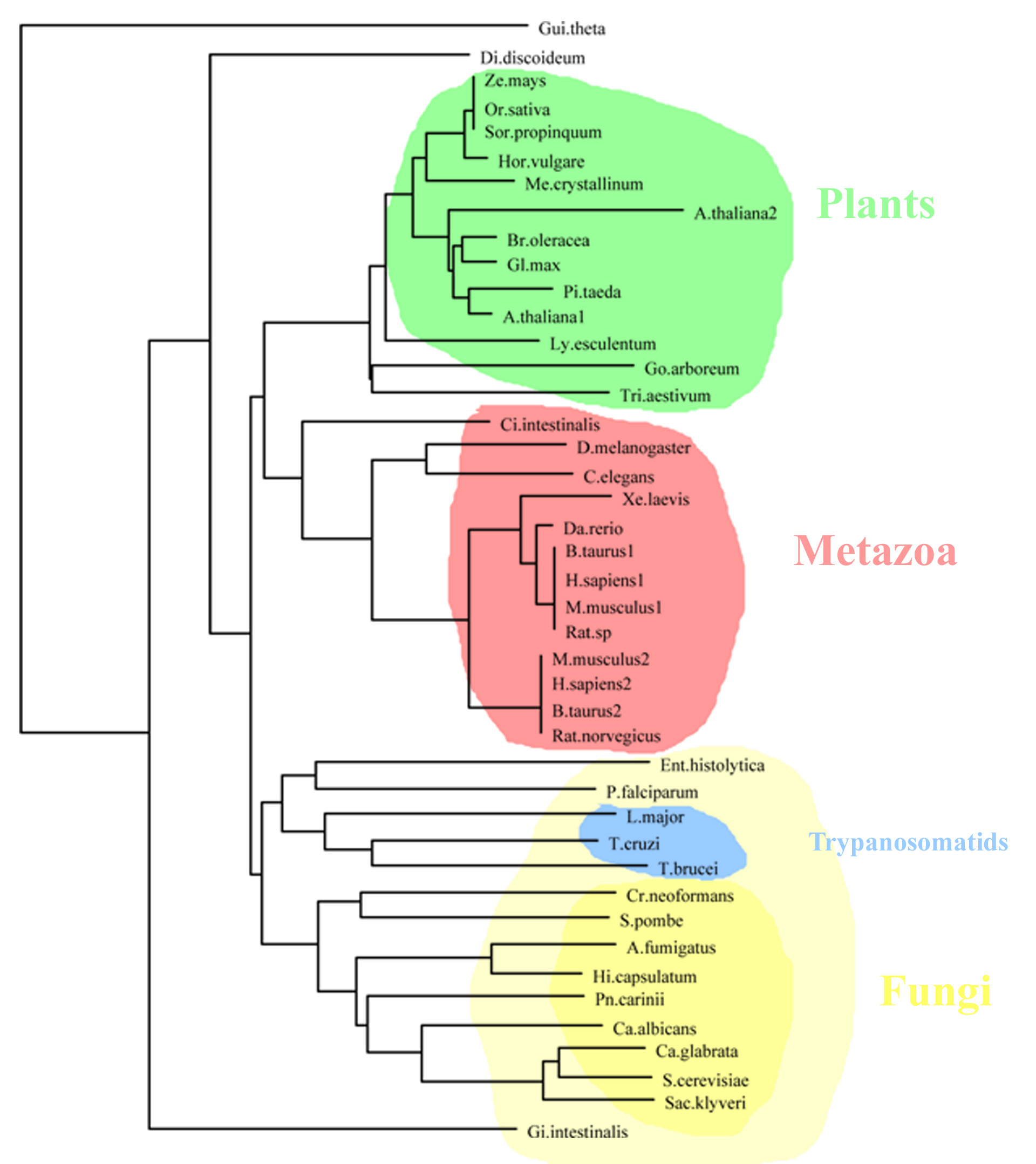
Most interestingly, we found that cluster separation becomes clearer when
analyzing the C-terminus, which is important in substrate recognition.
While the myristoylCoA binding site (N-terminus) had to remain highly
conserved (because of the single common ligand), distinct substrate
protein specificities (C-terminus) seem to have evolved dividing plants,
metazoa and fungi that cluster together with many other pathogenic
organisms, which, therefore, are potential targets for selective
inhibition of their NMTs.
Programs used: ClustalX (Thompson et al. 1997), PhyloDraw (Choi et al. 2000) and Phylowin (Galtier et al. 1996). Parameters: neighbor joining method; bootstrap with 1000 replicates; distance measure: observed divergence; alignment positions with gaps were excluded.
Accession numbers of the sequences:
Saccharomyces
cerevisiae NR P14743
Candida albicans NR P30418
Candida glabrata NR O74234
Schizosaccharomyces pombe NR O43010
Cryptococcus neoformans NR P34809
Histoplasma capsulatum NR P34763
Aspergillus fumigatus NR Q9UVX3
Arabidopsis thaliana1 NR AAF60968
Arabidopsis thaliana2 NR AAK49038
Leishmania major NR AAG38102
Plasmodium falciparum NR AAF18461
Drosophila melanogaster NR AAD27855
Caenorhabditis elegans NR P46548
Bos taurus1 NR P31717
Bos taurus2 NR Q9N181
Mus musculus2 NR O70311
Homo sapiens2 NR O60551
Mus musculus1 NR O70310
Homo sapiens1 NR P30419
Brassica oleracea NR AAF19802
Guillardia theta NR NP_113498
Amsacta moorei entomopoxvirus NR AAG02925
Melanoplus sanguinipes entomopoxvirus NR NP_048143
Giardia intestinalis HTGS AC075683(+1) AC041267(+3) AC088637(+1)
Saccharomyces klyveri GSS AZ124646(+2)
Trypanosoma brucei GSS AL490833(-3)
Trypanosoma cruzi GSS/EST AZ051049(+1) AI069625(+3)
Tetraodon nigroviridis GSS AL166863(+3) AL170798(+2) AL186230(-3)
AL319620(-2) AL319620(-3)
Saccharomyces servazzi GSS AL404522(+3)
Saccharomyces unisporus GSS AZ930605(+1)
Entamoeba histolytica GSS AZ672640(-3) AZ685745(+1)
Plasmodium vivax GSS AZ572682(-3)
Plasmodium berghei GSS AZ526668(+3) AZ525157(-1)
Danio rerio EST AW134126(+3) AI444250(+2) AI657581(+2) AI878266(+2)
Pleuronectes americanus EST AW013694(+1)
Sus scrofa EST F14725(+1)
Pneumocystis carinii EST AW334395(-1)
Aspergillus nidulans EST AI211114(+3)
Schistosoma mansoni EST AW017038(+2)
Rattus norvegicus EST BE126380(+3) C06728(+3) BF522952(+2) AI501085(-1)
Strongyloides stercoralis EST BE580365(+3)
Rattus sp EST AW141187(+1)
Meloidogyne javanica EST BG737070(+3)
Ciona intestinalis EST AV671484(+2) AV671935(+2) AV676318(-1)
Bombyx mori EST AV403090(+3)
Aspergillus niger EST BE759691(+1)
Triticum aestivum EST BE402458(+2) BE606254(+3) BE418143(+2)
Gallus gallus EST BG711647(+2)
Glycine max EST AW132453(+2) BF069724(+3) BF070843(+1)
Xenopus laevis EST AW645003(+1) BE026066(+3) AW766461(-1)
Medicago truncatula EST BF648024(+3) AW586887(+3)
Phytophthora infestans EST BE776171(+3)
Ceratopteris richardii EST BE641641(+3)
Pinus taeda EST AW982000(+2) AW290567(+1) AW888213(+3)
Oryza sativa HTGS AP003273(-1)
Zea mays EST BE644280(+1)
Sorghum propinquum EST BF481036(+2)
Hordeum vulgare EST BE194349(+2)
Dictyostelium discoideum EST AU060092(+2) AU034531(+3)
Gossypium hirsutum EST AI726036(+2)
Solanum tuberosum EST BE471614(+2) BG350597(+2)
Beta vulgaris EST BI095908(+2)
Lycopersicon esculentum EST AW649554(+3) AW224043(+2) AI897548(+1)
AW036027(+1) AW037713(+3)
Lotus japonicus EST AV409858(+1) AV423721(+3)
Mesembryanthemum crystallinum EST BE131169(-3)
Gossypium arboreum EST BF278989(+1) BF279264(+2) BG446339(+2)
Populus tremula EST BI069814(+1)
Sorghum bicolor EST BG465115(+2)