Supplementary material to:
TRENDS in Biochemical Sciences Vol.28 No.2 February 2003 p. 69-74
The
Tudor
Domain ‘Royal
Family’:
Tudor,
Plant
Agenet,
Chromo,
PWWP,
and
MBT Domains
Sebastian Maurer-Stroh1, Nicholas J. Dickens2, Luke Hughes-Davies3, Tony Kouzarides3, Frank Eisenhaber1 and Chris P. Ponting2*
1
Research Institute of Molecular Pathology, Dr. Bohr-Gasse 7, A-1030 Vienna,
Republic of Austria
2 MRC
Functional Genetics Unit, Department of Human Anatomy and
Genetics, University
of Oxford, South Parks Road, Oxford, OX1 3QX, UK
3
Cancer Research UK Laboratories, Wellcome Trust/Cancer Research UK
Institute,
University of Cambridge, Tennis Court Road, Cambridge, CB2 1QR, UK.
*Author for correspondence
Mail MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, South Parks Road, Oxford, OX1 3QX, UK
Tel. +44 (0)1865 272175
FAX +44 (0)1865 272420
E-mail Chris.Ponting@anat.ox.ac.uk
WWW http://www.smart.ox.ac.uk
Figure
S1a:
Alignment of Agenet domains
Figure S1b:
Alignment of Rsa1p with NUFIP and homologues
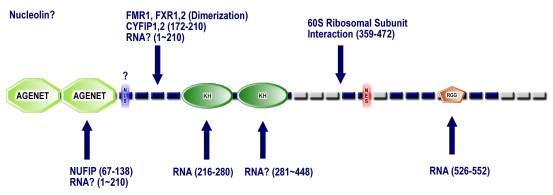
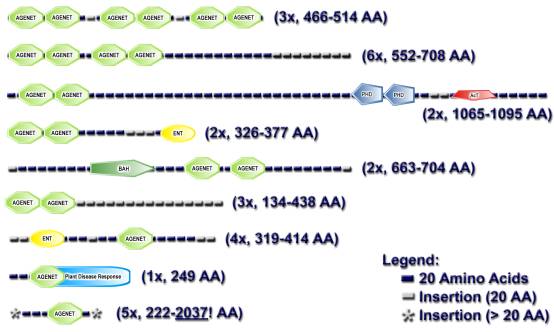
Figure S2: Domain
architecture of representative Agenet domain-containing proteins (A) and the
FMR1 protein (B)
62 Agenet domains were found in 28 different Arabidopsis thaliana proteins (Figure S1a). Homologues were detected in current sequence databases using an iterative combination of PSI-BLAST, employing an Expect (E)-value inclusion threshold of 5 x 10-3, and hidden Markov model (HMM) searches. Homologues were identified using PSI-BLAST, but their multiple alignment was achieved using HMMer.
We noted marginal, albeit non-significant, sequence similarity between NUFIP and a
Saccharomyces cerevisiae protein, Rsa1p. In order to investigate the hypothesis that NUFIP and Rsa1p are homologues, we identified the
Candida albicans Rsa1p orthologue in its incomplete genome (TBlastn search, E = 2 x 10-5). This employed a TBlastn search of incompletely-sequenced microbial genomes
(http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html).
Further Blastp, PSI-BLAST and TBlastn searches with this conceptual protein sequence demonstrated significant similarity between fungal Rsa1p and mammalian NUFIP sequences (E < 5 x 10-3) (Figure S1b).
As Rsa1p is involved in a nucleoplasmic assembly step of pre-60S ribosomal
subunits prior to translocation from the nucleus (Kressler et al., 1999 Mol.Cell.Biol.
19, 8633-8645), this observation provides further support for the
involvement of FMRP homologues in ribosome assembly (Siomi et al., 1996 Mol.Cell.Biol.
16, 3825-3832).
AT3G06520/167-236
DQYEKGALVEVRS(5)KGSWYCARILCLLG---DDKYIVEHLK(6)ESIPLRDVVE-AKDIRPVPPSEL
6xAG
AT3G06520/240-298
VCYEPGVIVDAWF---NKRWWTSRVSKVLGG-GSNKYSVFIIS-----TGEETTIL-NFNLRPHKDWIN 6xAG
AT3G06520/331-399
KVFNNGMEVEVRS(5)EASWFSAKIVSYLG---ENRYTVEYQT(5)ERELLKEEAR-GSDIRPPPPPLI
6xAG
AT3G06520/403-460
YRYELYELVDAWY---NEGWWSGRVYKINNN--KTRYGVYFQT-----TDESLEFA-YNDLRPCQVWRN 6xAG
AT1G09320/12-86
SYLKPGSAVEISS(5)RGSWYMGKVITIPS(5)SVKCQVEYTT(6)GTKPLKEVVD-MSQLRPPAPPMS 6xAG
AT1G09320/92-148
KKIVVGEEVDAFY---NDGWWEGDVTEVLD---DGKFSVFFRS-----SKEQIRFR-KDELRFHREWVD 6xAG
AT1G09320/424-480
SPFERHDKVNALY---NDGWWVGVIRKVLA---KSSYLVLFKN-----TQELLKFH-HSQLRLHQEWID 6xAG
AT3G62300/159-226
SDFSAGKSVEVRT(5)GDVWAPAMVIKEDE---DGTMLVKLKT(4)EVNCTKISVS-YSEIRPSPLPIG 4xAG
AT3G62300/228-284
RDYKLMENVDALV---ESGWCPGVVSKVLA---GKRYAVDLGP-----NRESKEFS-RLQLRPSIEWKD 4xAG
AT2G47230/78-134
IVLEEGTVVDADH---KDGWWTGVIIKKLE---NGKFWVYYDS-----PPDIIEFE-RNQLRPHLRWSG 4xAG
AT2G47230/223-279
EKYELMDRVEVFR---GSVWRQGLVRGVLD---HNCYMVCLVV-----TKEEPVVK-HSDLRPCKVWED 4xAG
AT4G32440/1-67
MRIRKGSRVEVFS(5)YGAWRCAEIVSGNG----HTYNVRFYS(4)HEEAVMEKVP-RKIIRPCPPLVD 2xAG
AT4G32440/69-125
ERWDTGELVEVLD---NFSWKAATVREELS---GHYYVVRLLG-----TPEELTFH-KVNLRARKSWQD 2xAG
AT4G17330/1571-1635
EDIKEGSNVEVFK(5)RTAWYSANVLSLED---DKAYVLFSDL---SVEQGTDKLKEWVALKGEGDQAP 2xAG
AT4G17330/1662-1720
HIWKIGDRVDSWV---HDSWLEGVITEKNKK-DENTVTVHFPA-----EEETLTIK-AWNLRPSLVWKD 2xAG
AT1G26540/6-77
MKITKDCVVEVSS(5)EGAWFRAVLEENPGNSSRRKLRVRYST(5)GSSPLIEHIE-QRFIRPVPPEEN 4xAG
AT5G23770/17-88
PMFSPGTMVEVSS(5)EVVWVPSMVIKEFKEDDEYKYIVKDKS(5)KKARPNKTVD-LSSLRPIPVSVD 2xAG
AT3G06520/77-136
RRFKADDEVDVFRDS-EGCWVRGNVTTVLE---DSRYIVEFKG----ENRPEIEVD-QFNLRLHREWLD 6xAG
AT1G11420/150-213
SVFSCGTMVELRF---DCAWIPVIVIKELEK--DKRFLVKYWN(4)CRESKNLIVD-SLRLRPMQPPLS 4xAG
AT1G06340/1-71
MEFVKGDQVEVCS(5)LGSYFGATVVSKTPE--GSYYKIKYKN(6)QSKRLVEVIS-ADELRPMPPKSL
2xAG
AT5G20030/64-120
DAWCPGDILEVFQ---SCSWKMAIVSKVLG---NGCFLVRLLG-----SSLKFKVT-KSDIRVRQSWQD 2xAG
AT5G23800/10-78
LSLSEGCEVEISY(5)ESVWYKAILEAKPNSIFKEELSVRLLKD-DFSTPLNELRH-KVLIRPIPPTNV 4xAG
AT5G55600/384-442
FDLTIGEAVDAWW---NDGWWEGVVIATGKP-DTEDLKIYIPG-----ENLCLTVL-RKDIRISRDWVG AG
AT5G52070/55-115
DSWKVGDLVDWLR---DDIYWSGEIVEMRG---RRACQIELLP-KPEGEGDSYQGL-CKNLRPRLDWSV AG
AT5G58610/105-162
LNLAYGLCVDVFF---SDAWWEGVLFDHEN--GSEKRRVFFPD-----LGDELDAD-LQSLRITQDWNE
AG,2xPHD,GNAT
AT1G68580/361-427
HHIKKGSLIEVLS(5)RGCWFKALVLKKHK----DKVKVQYQD(4)DDESKKLEEW-ILTSRVAAGDHL
BAH,2xAG
AT3G12140/133-192
AEALIGRKVWTKWPE-DNHFYEAIITQYNA--DEGRHALVYDI-----HAANETWE-WVDLKEIPPEDI
AG
AT3G57970/198-259
PGSLVGRRVHIQMPD-EDEYIEFLITKYDAN--TETHHLLSAF---SNKDYEDPCN-WVDLRHVQAEDM AG
AT1G02740/50-117
GHFEEGERVLAKH---SDCFYEAKVLKVEFKDNEWKYFVHYIV(5)NIEKQKEQGLKQQGIKSAMAWKV AG
Consensus/75%
..bp.Gp.V-sb....pssWb.t.l.p......p.pb.Vbb.s.....p..pbphp.b.plRs..s..s
2-structure
EEEEE
eEEEEEEEe EEEEEEe
ee
Figure legend S1a:
Multiple sequence alignment of plant Agenet domain sequences represented using
CHROMA and a 75% consensus threshold. 29 representatives of the 62
Arabidopsis thaliana Agenet domain family. Sequences have been chosen that are less than 40% identical to any other homologue. Secondary structures
predicted (*)
at expected accuracies of > 82 % (E) or >72% (e) are indicated below the alignment (E/e, extended or b-strand structure). The domain contents of these sequences are shown following the alignment: Ag, Agenet domains; BAH, Bromo adjacent homology domains; GNAT, GCN5-related
N-acetyltransferase domain; and, PHD, plant homeodomains.
The GenInfo identifiers for these gene products are: AT3G06520 (15230734), AT1G09320 (15217483), AT3G62300 (15228725), AT2G47230 (15226533), AT4G32440 (15236804), AT4G17330 (15236041), AT1G26540 (15222723), AT5G23770 (15237825), AT1G11420 (6554201), AT1G06340 (15221452), AT5G20030 (15241247), AT5G23800 (15237832), AT5G55600 (18087548), AT5G52070 (15242269), AT5G58610 (15237720), AT1G68580 (15221440), AT3G12140 (9294111), AT3G57970 (15230909), and AT1G02740 (15217854).
RSA1p
Sc IALITDEDVKKWREERKKMW--LLKISNNK
234-261 (6325063)
RSA1p
Ca ISLQTEEDIEKWIEERKRNWPTNKNIELKR
SPBC16C6.03c
Sp ISINTPEEIEAWIQERKKNWPTESNIRSKQ 97-126
(7493660)
RSA1p
Af STLQSPTDIAAWIEERKKRFPTQAKAEEKR
NUFIP
Hs IKLDTPEEIARWREERRKNYPTLANIERKK 228-253 (11433762)
NUFIP
Pf IILNDAKEIEKWISERKKNYPTRNKILNNM
CG4076
Dm KKVWSEEELAAWRAERRKKFPTAANVELAR 140-169 (7293869)
At5g18440
At ALMYTPREVQQWREARRKNYPTKFLVEKKV 236-265
(15238815)
EST
Hv PIIYDKNEVKQWVQARKKNYPTRANVNKKL
(14525941)
Consensus/75%
..l.*.c-l..WbpER+KpaPTb.plpppb
Figure legend S1b:
Multiple sequence alignment of the most highly-conserved region of Rsa1p/NUFIP homologues, represented using CHROMA and a 75% consensus threshold. The Plasmodium falciparum NUFIP1 was found on chromosome 14 using http://tigrblast.tigr.org/euk-blast/index.cgi?project=pfal. Amino acid numbers and GenInfo identifiers (if known) are shown following the alignment. Species: Af, Aspergillus fumigatus; At, Arabidopsis thaliana; Ca, Candida albicans; Dm, Drosophila melanogaster; Hs, Homo sapiens; Hv, Hordeum vulgare; and, Pf, Plasmodium falciparum.
Schematic representation of the domain architectures of (A) a representative set of Agenet domain-containing proteins, and (B) the FMR1 protein.
A
B