

 |
GPI Lipid Anchor Project |  |


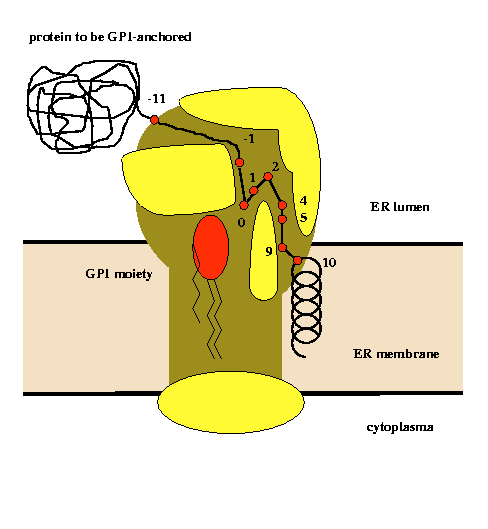
Proprotein sequences to be posttranslationally GPI-modified are searched for in complete genomes with the recently developed big-PI predictor. The results indicate that GPI-anchoring is present among Eukaryota and possibly among a subgroup of the Archaea but it appears absent in other Archaea and all Eubacteria studied. The chromosomal distribution of GPI-modified proteins is uneven. Dozens of proteins annotated just as hypothetical could be characterized as GPI-anchored.

|
An analysis of physical properties of amino acid residues at given sequence positions in the vicinity of the GPI-modification site allowed the construction of a model of the active site of the putative transamidase complex (Eisenhaber et al., Prot.Eng., 11, 1155-1161,1998). |


A.thaliana:
ftp://ncbi.nlm.nih.gov/genbank/genomes/A_thaliana/
C.elegans:
ftp://ftp.sanger.ac.uk/pub/databases/wormpep/wormpep17
ftp://ftp.sanger.ac.uk/pub/databases/wormpep/wormpep31
L.major:
http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/altik?gi=13580&db=Genome&from=0&to=0
P.falciparum:
Chr.02: ftp://ftp.tigr.org/pub/data/p_falciparum/chr2/complete_sequence/
Chr.03: ftp://ncbi.nlm.nih.gov/genbank/genomes/P_falciparum/
S.cerevisiae:
ftp://ncbi.nlm.nih.gov/genbank/genomes/S_cerevisiae
All complete genomes for Archaea and Eubacteria:
ftp://ncbi.nlm.nih.gov/genbank/genomes/bacteria
with the exception of
Pyrococcus furiosus:
http://www.genome.utah.edu
Campylobacter jejuni:
ftp://ftp.sanger.ac.uk/pub/pathogens/cj
Chlamydia trachomatis MoPn:
ftp://ftp.tigr.org/pub/data/c_trachomatis/
Helicobacter pylori:
ftp://ftp.tigr.org/pub/data/h_pylori

Last modified: 13th June 2002