|
Supplementary material for the publication:
"The STIR domain superfamily (SEFIR, TIR)
in signal transduction, development and immunity"
Trends Biochem Sci. 2003 May;28(5):226-9, PMID: 12765832
Novatchkova M, Leibbrandt A, Werzowa J,
Neubüser A and Eisenhaber F
|

|
|
|

|
|
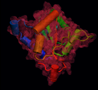
Collection of some SE/IR family using the full chicken SEF sequence as a starting point.
Simple BLAST search of full length cSEF against the NRDB.
|
|
Collection of the STIR superfamily using chicken SEF as a starting point.
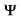 -BLAST search of chicken SEF (310-450) against the NRDB. -BLAST search of chicken SEF (310-450) against the NRDB.
- round 1
- round 2
- round 3
-> This search reveals a weak similarity of the proximal region of chicken SEF (residues 310-450) to small bacterial proteins with a N-terminal TIR domain (BAB83481 and NP_662351). The similarities of the SEF/IL17R membrane proximal domain to these proteins is below conventionally used PSI-BLAST thresholds and are thus indicative of homology, but the significance has to be supported further. A reciprocal psi-blast allowed us to support the similarity of BAB83481 (aligns to 80.7% of the CD TIR domain) to SEFIR proteins and to collect the STIR family.
|
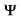 -BLAST search of BAB83481 (1-150) against the NRDB. -BLAST search of BAB83481 (1-150) against the NRDB.
|
|

|
|
The STIR SUPERFAMILY was collected using saturated blast starting with the residues 1-150 from BAB83481.
The STIR protein family is represented by
bacterial (fa)
plantal (fa)
and metazoan (fa) sequences in NRDB.
The STIR SUPERFAMILY protein set (fasta with too short sequences being excluded) was used to build a multiple sequence alignment (pdf).
A protein set made non-redundant with a cut off of 50% identity was used to construct a HMM.
A subset including SEFIR proteins only was used to construct a SEFIR specific HMM.
|
|

|
 Chicken SEF (cSEF) was isolated in a screen for FGF4/8 regulated genes in chicken facial mesenchyme. A summary on the initial experimental functional analysis of chicken SEF is published on the web. Chicken SEF (cSEF) was isolated in a screen for FGF4/8 regulated genes in chicken facial mesenchyme. A summary on the initial experimental functional analysis of chicken SEF is published on the web.
|
|